Fellowship Profile Series 2: Jessica Orozco, 2016 Indigenous Ethnobiologist Fellow
Fellowship Profile Series 2: Jessica Orozco, 2016 Indigenous Ethnobiologist Fellow
Words: Jessica Orozco
Since receiving the Society of Ethnobiology’s Indigenous Ethnobiologist Fellowship I have conducted a preliminary survey of Desert Chia’s (Saliva columbariae) genetic variation that will be used to guide further research into the evolution and biology of this ubiquitous species. Salvia columbariae is an herbaceous annual in the Mint family (Lamiaceae) that grows in a wide range of environments throughout California and the southwestern United States, including Sonora, Mexico. It has also been prized for its highly nutritious seed, which was utilized and traded by Native Americans throughout its range. Seeds were transported by Indigenous peoples deliberately, but were also likely dropped or discarded along native travel routes, contributing to the spread of this species into new areas as well.
 As a generalist species, S. columbariae is able to thrive in a wide variety of environmental conditions. Salvia columbariae has a wide geographic distribution and can be found growing on a variety of substrates. While herbarium specimens vary widely in morphology, this is believed/understood to be due to phenotypic plasticity, although there have not been any common garden studies to prove this. I am ultimately interested in seeing if the variation in geographic distribution and phenotypic plasticity translates to genetic variation and the formation of genetically distinct groups. The trading of S. columbariae seeds among Native American tribes could have contributed to its current distribution while also introducing admixture into naturally occurring populations, altering the genetic structure of populations.
As a generalist species, S. columbariae is able to thrive in a wide variety of environmental conditions. Salvia columbariae has a wide geographic distribution and can be found growing on a variety of substrates. While herbarium specimens vary widely in morphology, this is believed/understood to be due to phenotypic plasticity, although there have not been any common garden studies to prove this. I am ultimately interested in seeing if the variation in geographic distribution and phenotypic plasticity translates to genetic variation and the formation of genetically distinct groups. The trading of S. columbariae seeds among Native American tribes could have contributed to its current distribution while also introducing admixture into naturally occurring populations, altering the genetic structure of populations.
In this study, I used phylogenetic methods to assess genetic variation in S. columbariae across its range. I extracted DNA from 16 herbarium specimens of S. columbariae from Rancho Santa Ana Botanic Garden’s herbarium to represent S. columbariae across its range. I also used Genbank accessions for 13 out-group taxa from Audibertia (the section to which S. columbariae belongs and includes: Salvia dorrii var. dorrii, S. dorrii var. mearnsii S. dorrii var pilosa, S. pachyphylla, S. leucophylla, S. mellifera, S. mohavenisis, S. Munzii, S. sonomensis and S. greatae). Three taxa from outside of Audibertia (S. carnea from the subgenus Calospace, Rosmarinus officinalis, and Meriandra bengalensis) were used to help root the tree and to place S. columbariae with the Audibertia clade.
Phylogenetic trees were constructed using Sank off parsimony, Bayesian inference (BI) and Maximum Likelihood (ML) approaches. Prior to this study, there were no data available in Genbank for S. columbariae, and so a goal of this study was to confirm the placement of S. columbariae in this group. The nuclear ribosomal ITS region was used to assess genetic variance in S. columbariae, as well as the section Audibertia. Based on the evidence produced in this study, S. columbariae forms a monophyletic group within the Audibertia clade. The study was able to identify some populations that formed geographic clades, but with low support values. These derived clades include populations from the Sonoran desert region and from Lake Country, CA. Given the current data, these two clades represent the far reaches of S. columbariae's distribution, which leads one to believe that there might be some kind of relationship between geography and population lineage.
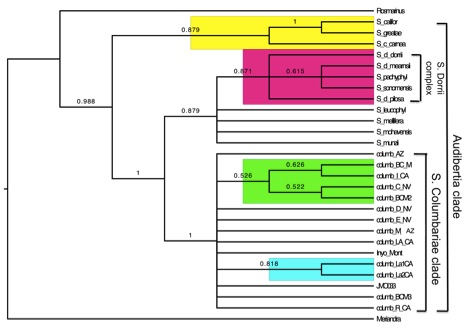
The phylogenetic relationships within S. columbariae clade show some genetic structure using the ITS marker, but in order to accuraely identify distinct genotypes or introgressed loci, a large number of nuclear regions, rather than high-copy nuclear ribosomal markers are required. Future research goals would be to include a genotyping study using single nucleotide polymorphisms (SNPs) to measure the amount of genetic variation between populations.
